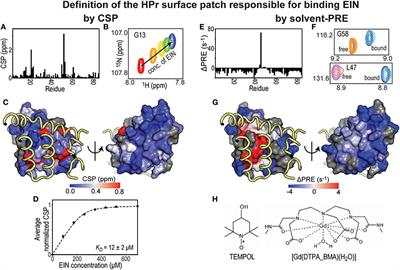
Atomic resolution protein allostery from the multi-state structure of a PDZ domain | Nature Communications

Systematic Exploration of Protein Conformational Space Using a Distance Geometry Approach | Journal of Chemical Information and Modeling
![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/4-Table3-1.png)
PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar
Determination of three-dimensional structures of proteins in solution by nuclear magnetic resonance spectroscopy

Computational prediction of protein–protein binding affinities - Siebenmorgen - 2020 - WIREs Computational Molecular Science - Wiley Online Library

Quantitative Interpretation of Solvent Paramagnetic Relaxation for Probing Protein–Cosolute Interactions | Journal of the American Chemical Society

Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin | Science Advances
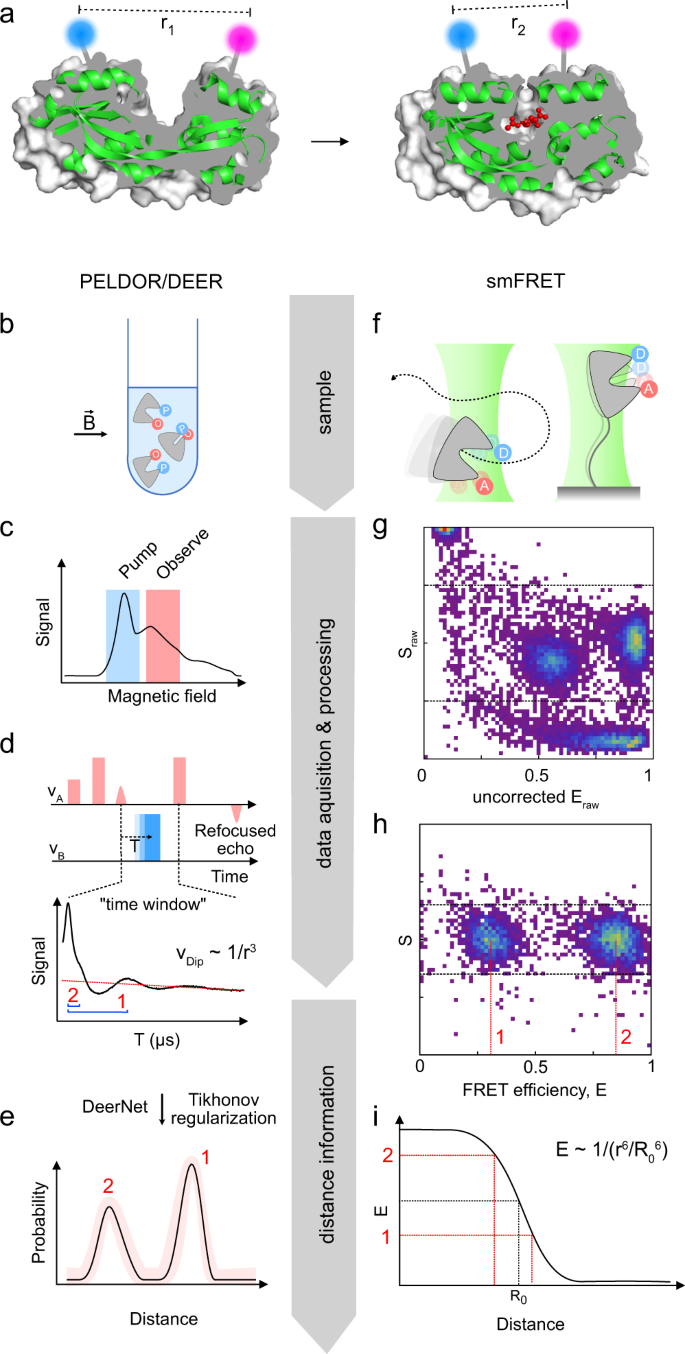
Cross-validation of distance measurements in proteins by PELDOR/DEER and single-molecule FRET | Nature Communications

Frontiers | An Integrative Approach to Determine 3D Protein Structures Using Sparse Paramagnetic NMR Data and Physical Modeling

Quantitative Interpretation of Solvent Paramagnetic Relaxation for Probing Protein–Cosolute Interactions | Journal of the American Chemical Society

Computational generation of proteins with predetermined three-dimensional shapes using ProteinSolver - ScienceDirect
![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/3-Table2-1.png)
PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar